Standard PCR Protocol

Polymerase Chain Reaction (PCR) Protocol
Introduction
Polymerase Chain Reaction (PCR) is a cornerstone technique in molecular biology, enabling the amplification of specific DNA sequences from minute amounts of starting material. Since its invention by Kary Mullis in 1983, PCR has revolutionized genetic research, diagnostics, forensics, and biotechnology. This protocol provides a comprehensive guide to performing standard PCR, including detailed steps, reagent specifications, troubleshooting, and optimization strategies.
Principle of PCR
PCR is an enzymatic process that amplifies a specific DNA segment through repeated cycles of denaturation, annealing, and extension. The reaction relies on a thermostable DNA polymerase, typically Taq polymerase, which withstands high temperatures required to denature double-stranded DNA (Figure 1).
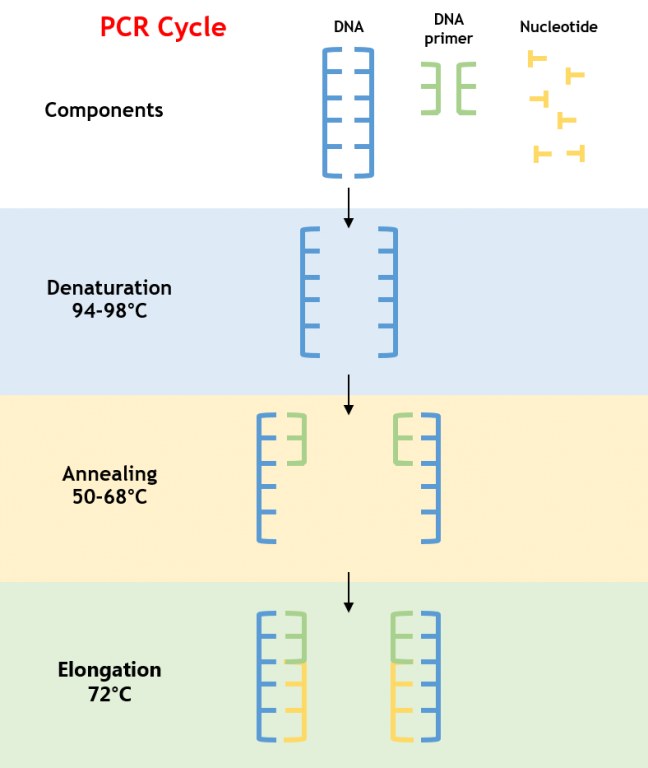
Figure 1: PCR Amplification Process
- Denaturation: DNA is heated to 94–98°C, breaking hydrogen bonds to yield single-stranded DNA.
- Annealing: The temperature is lowered to 50–68°C, allowing primers to bind to complementary sequences flanking the target DNA.
- Extension: The temperature is raised to 72°C, optimal for Taq polymerase to synthesize new DNA strands by adding deoxynucleotide triphosphates (dNTPs).
Materials and Equipment
Before starting, ensure all reagents and equipment are available and properly stored. Use molecular biology-grade reagents to avoid contamination or inhibition.
Reagents
- Taq DNA Polymerase: Stored at -20°C.
- 10X PCR Buffer: Typically includes 100 mM Tris-HCl, 500 mM KCl, and 15 mM MgCl₂ (pH 8.3–8.8). Some buffers lack MgCl₂, requiring separate addition. Store at -20°C or 4°C per manufacturer.
- Magnesium Chloride: 25 mM MgCl₂ Solution (if not included in PCR buffer). Store at -20°C or 4°C.
- dNTPs: Deoxynucleotide triphosphates (dATP, dTTP, dCTP, dGTP; 10 mM each, combined or separate). Store at -20°C.
- Primers: Forward and reverse primers (100 µM stock in TE buffer, diluted to 10 µM working solution).
- Template DNA: Genomic DNA (10–100 ng), plasmid DNA (0.1–1 ng), or cDNA (1–10 ng), depending on the application. Store at -20°C.
- Nuclease-Free Water: Molecular biology-grade, sterile water. Store at room temperature.
- Master Mix
- SYBR® Safe DNA Gel Stain: For agarose gel visualization (alternative: ethidium bromide, but use with caution due to toxicity). Store at 4°C.
- 6X Loading Dye: For gel electrophoresis (e.g., containing bromophenol blue and xylene cyanol). Store at 4°C.
- DNA Ladder: 100 bp or 1 kb ladder, depending on expected amplicon size. Store at -20°C.
Equipment
- Thermal Cycler: Programmable PCR machine.
- PCR Tubes: 0.2 mL thin-walled PCR tubes or 96-well plates, compatible with the thermal cycler.
- Pipettes and Tips: Calibrated pipettes (P2, P10, P20, P200, P1000) with filter tips to prevent contamination.
- Microcentrifuge: For brief spins to collect reaction components.
- Agarose Gel Electrophoresis System: Including gel casting tray, comb, power supply, and UV transilluminator (or blue light for SYBR Safe).
- Vortex Mixer: For mixing reagents.
- Ice Bucket: To keep reagents cold during setup.
- Spectrophotometer: For quantifying DNA concentration.
PCR Protocol
Step 1: Primer Design and Preparation
Primers are short, single-stranded DNA oligonucleotides (18–25 bases) that flank the target sequence and serve as starting points for DNA synthesis. Proper primer design is critical for specific and efficient amplification.
Primer Design Guidelines
Use software tools to design primers. Follow these criteria:
- Length: 18–25 bases. Shorter primers (<18) may lack specificity; longer primers (>25) may form secondary structures.
- Melting Temperature (Tm): 55–65°C, with forward and reverse primers within 2°C of each other. Calculate Tm using the formula for a basic estimate, or use software for accuracy (accounts for salt and primer concentration).
Tm = 4 (G+C) + 2 (A+T)
- GC Content: 40–60%. Avoid extremes (<30% or >70%) to prevent poor annealing or secondary structures.
- 3' End: End with a G or C (stronger base pairing) and avoid runs of three or more identical bases (e.g., GGG) to reduce mispriming.
- Specificity: Check for specificity using NCBI BLAST (blast.ncbi.nlm.nih.gov) to ensure primers bind only the target sequence. Avoid homology to non-target regions.
- Secondary Structures: Avoid hairpins, self-dimers, and hetero-dimers. Delta G for dimers should be >-5 kcal/mol.
- Amplicon Size: Design for 100–1000 bp amplicons for standard PCR. Longer amplicons (up to 5 kb) are possible but require optimization.
- Avoid Repeats: Do not place primers in regions with repetitive sequences or low complexity (e.g., ATATAT).
Primer Synthesis and Storage
- Ordering: Order HPLC-purified primers. HPLC purification ensures high purity, reducing non-specific amplification.
- Resuspension: Resuspend lyophilized primers in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) to a 100 µM stock concentration.
- Working Solution: Dilute stock to 10 µM in nuclease-free water. Store at -20°C.
- Storage: Store 100 µM stocks at -20°C for up to 1 year; 10 µM working solutions are stable at -20°C for 6 months or 4°C for 1 month. Avoid repeated freeze-thaw cycles (>10) to prevent degradation.
Primer Quality Control
- Quantification: Verify primer concentration using a spectrophotometer (A260). Expected absorbance for 100 µM is ~0.3–0.5, depending on sequence.
- Integrity: Optional—run primers on a 2% agarose gel to confirm no degradation (should appear as a single, sharp band).
- Test Amplification: Perform a trial PCR with a known template to confirm primer functionality before large-scale experiments.
Step 2: Reaction Setup
The PCR reaction mixture contains all components necessary for DNA amplification. The provided protocol is scaled to a 50 µL reaction volume, with a master mix for 11 reactions (10 samples + 1 no-template control) to account for pipetting losses.
Reaction Components
The table below lists reagents and volumes for one 50 µL reaction and the master mix for 11 reactions.
| Reagent | Volume per Reaction (µL) | Total for 11 Reactions (µL) |
| Nuclease-Free Water | 36.5 | 401.5 |
| 10X PCR Buffer | 5.0 | 55.0 |
| dNTPs (10 mM each) | 1.0 | 11.0 |
| MgCl₂ (25 mM) | 3.0 | 33.0 |
| Forward Primer (10 µM) | 1.0 | 11.0 |
| Reverse Primer (10 µM) | 1.0 | 11.0 |
| Taq Polymerase (5 U/µL) | 0.5 | 5.5 |
| Template DNA (variable) | 3.0 | Add separately |
Step 3: Thermal Cycling
The thermal cycler subjects the reaction to repeated temperature changes to facilitate denaturation, annealing, and extension. The provided cycling conditions are optimized for standard PCR with Taq polymerase and amplicons of 100–1000 bp.
|
|
|
|
|
NeoCycler Trio - Multi-block Thermal cycler (Cat# NB-12-3024) NeoCycler Duo - Multi-block Thermal cycler (Cat# NB-12-3025)
|
NeoCycler 300 - Fast Gradient Thermal Cycler (with 9677 / 96 module) (Cat# NB-12-3001) NeoCycler 200 - Gradient Thermal Cycler (Cat# NB-12-3002) NeoCycler 100 - Non-Gradient Thermal Cycler (with 9677 / 96 module) (Cat# NB-12-3003) |
|
|
|
|
|
|
NeoCycler 600 - Super Gradient Thermal Cycler (Cat# NB-12-3026) |
NeoCycler Mini (Cat# NB-12-3029-2) |
Thermal Cycling Program
Program the thermal cycler with the following steps:
- Initial Denaturation: 95°C for 2 minutes
- Denatures double-stranded template DNA and activates Taq polymerase (hot-start versions may require longer, e.g., 5–10 minutes).
- Cycling (30–35 cycles):
- Denaturation: 95°C for 30 seconds
- Separates DNA strands. Shorter times (15–20 seconds) may suffice for small templates (e.g., plasmids).
- Annealing: 55–65°C for 30 seconds
- Primers bind to complementary sequences. Set 2–5°C below the lower primer Tm (e.g., if Tm is 60°C, use 55–58°C). Optimize if needed (see Section 8).
- Extension: 72°C for 1 minute per kb
- Taq polymerase synthesizes new DNA. For a 500 bp amplicon, use 30–60 seconds; for 1 kb, use 60 seconds. Minimum 30 seconds for <500 bp.
- Denaturation: 95°C for 30 seconds
- Final Extension: 72°C for 5 minutes
- Completes partial extensions and adds 3' A-overhangs (useful for TA cloning).
- Hold: 4°C indefinitely
- Stores reactions until retrieval. Avoid prolonged storage (>24 hours) to prevent degradation.
Cycle Number
- Standard: 30 cycles for moderate template amounts (10–50 ng genomic DNA).
- Low Template: 35 cycles for low template amounts (<1 ng) or cDNA.
- High Template: 25–28 cycles for high template amounts (>100 ng) to reduce non-specific products.
- Excess Cycles: >35 cycles may increase non-specific amplification or primer dimers.
Thermal Cycler Settings
- Lid Temperature: Set to 105°C to prevent condensation.
- Ramp Rate: Use default settings (2–5°C/second). Faster ramps may require adjustment for older machines.
- Volume: Set to 50 µL in the cycler program for accurate heating.
Variations
- Short Amplicons (<200 bp): Reduce extension time to 15–30 seconds.
- Long Amplicons (1–5 kb): Increase extension time (1–2 minutes/kb) and consider high-fidelity polymerases.
- GC-Rich Templates: Use higher denaturation temperatures (98°C) or additives like DMSO (5–10%).
Step 4: Analysis of PCR Products
Post-PCR, analyze products to confirm amplification success and amplicon size. Agarose gel electrophoresis is the standard method.
Agarose Gel Preparation
- Gel Concentration:
- 1% agarose for 500–5000 bp amplicons.
- 1.5–2% agarose for 100–500 bp amplicons.
- Prepare Gel:
- Weigh agarose.
- Dissolve in 1X TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) by microwaving until clear.
- Cool to ~50°C, add SYBR Safe (1 µL per 10 mL gel, per manufacturer), and pour into a casting tray with a comb.
- Let solidify for 20–30 minutes.
- Setup: Place gel in an electrophoresis tank filled with 1X TAE buffer.
Sample Preparation
- Mix Sample: Combine 10 µL PCR product with 2 µL 6X loading dye (final 1X).
- Load Ladder: Load 5–10 µL DNA ladder (100 bp or 1 kb, depending on amplicon size) in one lane.
- Load Samples: Load 12 µL of each PCR product + dye mix into separate wells. Include the No-Template Control (NTC) to check for contamination.
Electrophoresis
- Run Gel: Run at 80–120 V for 30–60 minutes, until the loading dye (bromophenol blue) migrates ~2/3 of the gel length.
- Visualize: Use a UV transilluminator (or blue light for SYBR Safe) to visualize bands. Photograph the gel for documentation.
Interpretation
- Expected Band: Compare band size to the DNA ladder. It should match the predicted amplicon size (based on primer design).
- NTC: No bands should appear in the NTC lane. Bands indicate contamination or primer dimers.
- Smearing: Indicates non-specific amplification, degraded template, or excess DNA.
- No Bands: Suggests failed amplification.
Post-Analysis
- Storage: Store PCR products at -20°C for weeks or 4°C for 1–2 days.
- Purification: For downstream applications (e.g., sequencing, cloning), purify products using a PCR cleanup kit or gel extraction kit.
- Quantification: Measure product concentration using a Nanodrop or fluorometer if needed.
Quality Control and Validation
To ensure reliable results, incorporate quality control at every step.
Template Quality
- Purity: A260/A280 ratio of 1.8–2.0 indicates pure DNA. Lower ratios suggest protein or RNA contamination.
- Integrity: Run template DNA on a 0.8% agarose gel to confirm no degradation (genomic DNA should appear as a high-molecular-weight band).
- Concentration: Use 10–100 ng genomic DNA or 0.1–1 ng plasmid DNA per 50 µL reaction.
Reagent Quality
- dNTPs: Check for degradation (store at -20°C, avoid >10 freeze-thaw cycles).
- Taq Polymerase: Verify activity by testing with a known template-primer pair.
- Primers: Confirm concentration and integrity.
Controls
- NTC: Detects contamination or primer dimers.
- Positive Control: Use a known template-primer pair to confirm reaction conditions.
- Negative Control (optional): Omit polymerase to check for non-specific amplification.
Gel Analysis
- Ladder: Ensure the ladder resolves clearly to confirm gel quality.
- Band Intensity: Strong, single bands indicate successful amplification. Weak or multiple bands suggest optimization is needed.
Troubleshooting
PCR may require optimization to achieve specific, high-yield amplification. Below are common issues and solutions.
No Amplification
- Cause: Low template concentration, degraded template, incorrect primers, or suboptimal annealing temperature.
- Solutions:
- Increase template (up to 200 ng) or cycles (up to 35).
- Verify template integrity on a gel.
- Confirm primer sequences and Tm; redesign if necessary.
- Perform a gradient PCR (50–65°C annealing) to find optimal temperature.
- Check Taq polymerase activity with a positive control.
Non-Specific Bands
- Cause: Low annealing temperature, high MgCl₂, excess primers, or non-specific primer binding.
- Solutions:
- Increase annealing temperature by 1–2°C increments.
- Reduce MgCl₂ to 1–1.2 mM.
- Decrease primer concentration to 0.1–0.15 µM.
- Redesign primers for higher specificity (use BLAST).
- Reduce cycle number to 25–28.
Smearing
- Cause: Excess template, degraded template, or over-cycling.
- Solutions:
- Reduce template to 10–50 ng.
- Verify template integrity.
- Decrease cycles to 25–30.
- Use fresh dNTPs and polymerase.
Primer Dimers
- Cause: Primers forming self- or hetero-dimers, visible as low-molecular-weight bands (<100 bp) in NTC.
- Solutions:
- Redesign primers to minimize dimer formation.
- Reduce primer concentration to 0.1 µM.
- Increase annealing temperature.
- Use hot-start Taq polymerase to prevent non-specific priming during setup.
Optimization Strategies
- Gradient PCR: Test a range of annealing temperatures (e.g., 50–65°C) in one run if the cycler supports gradient function.
- MgCl₂ Titration: Test 1–4 mM MgCl₂ in 0.5 mM increments.
- Additives: For GC-rich templates, add 5–10% DMSO or betaine (1–2 M) to the reaction.
- Hot-Start PCR: Use hot-start Taq (e.g., Platinum Taq) to reduce non-specific amplification during setup.
- Touchdown PCR: Start annealing at 5–10°C above Tm, decreasing 0.5°C per cycle for 10 cycles, then continue at the lower temperature. Enhances specificity.
Conclusion
This comprehensive PCR protocol provides a robust framework for amplifying DNA with high specificity and yield. By following the detailed steps for primer design, reaction setup, thermal cycling, and product analysis, users can achieve reliable results for a wide range of applications. Optimization and troubleshooting tips ensure success even with challenging templates or primers. For advanced applications, adapt the protocol as described, and consult manufacturer guidelines for specialized reagents or equipment.





